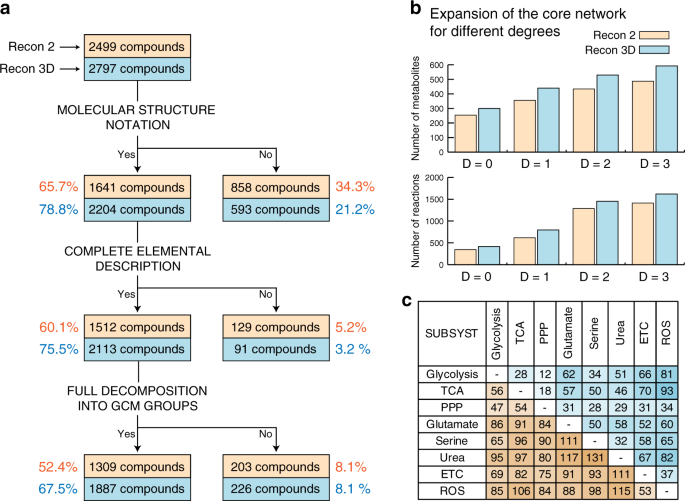
Recon3D enables a three-dimensional view of gene variation in human metabolism | Nature Biotechnology

Cofactors revisited – predicting the impact of flavoprotein-related diseases on a genome scale | bioRxiv
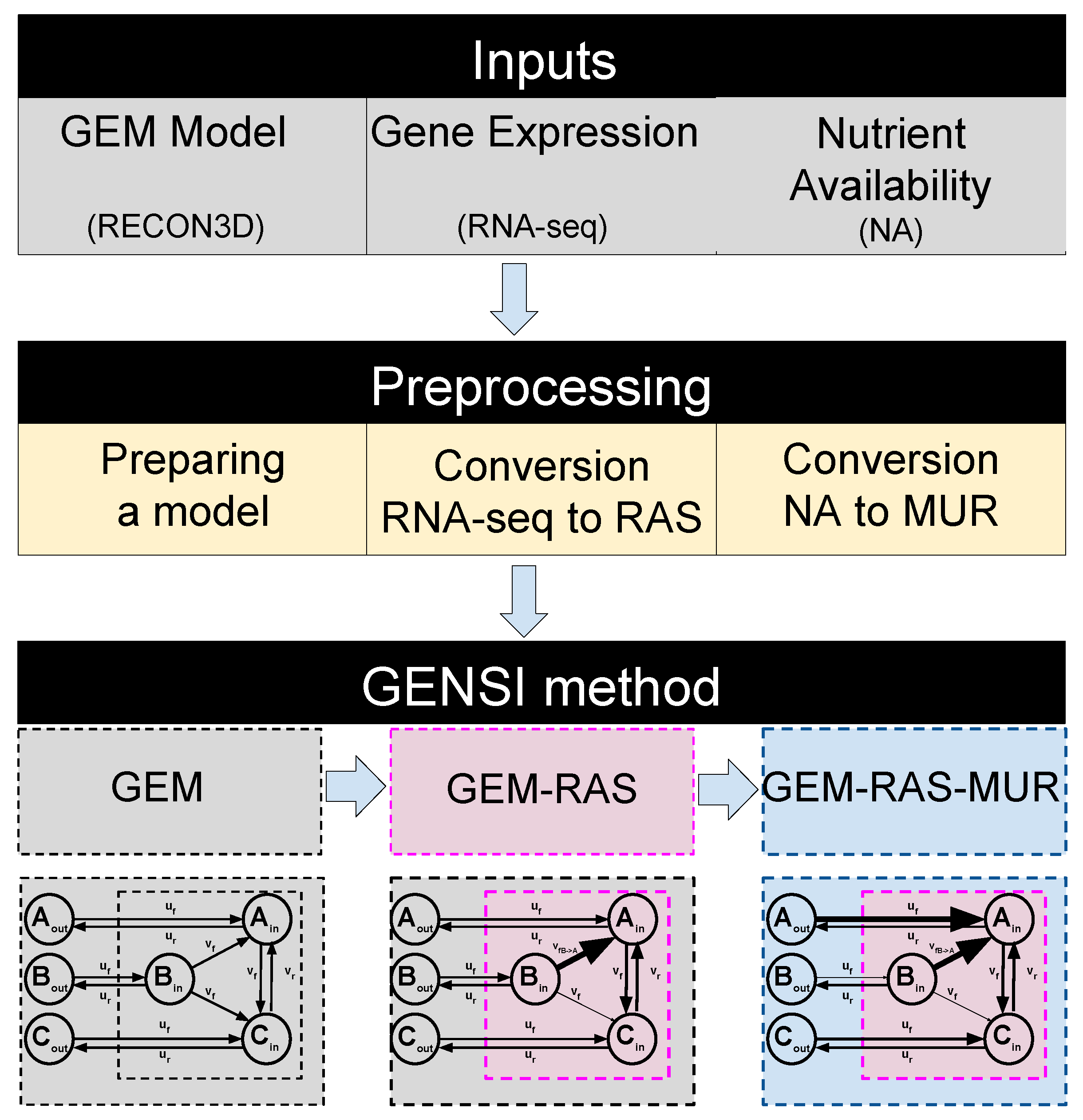
Biomolecules | Free Full-Text | Simultaneous Integration of Gene Expression and Nutrient Availability for Studying the Metabolism of Hepatocellular Carcinoma Cell Lines | HTML

Recon3D enables a three-dimensional view of gene variation in human metabolism | Nature Biotechnology

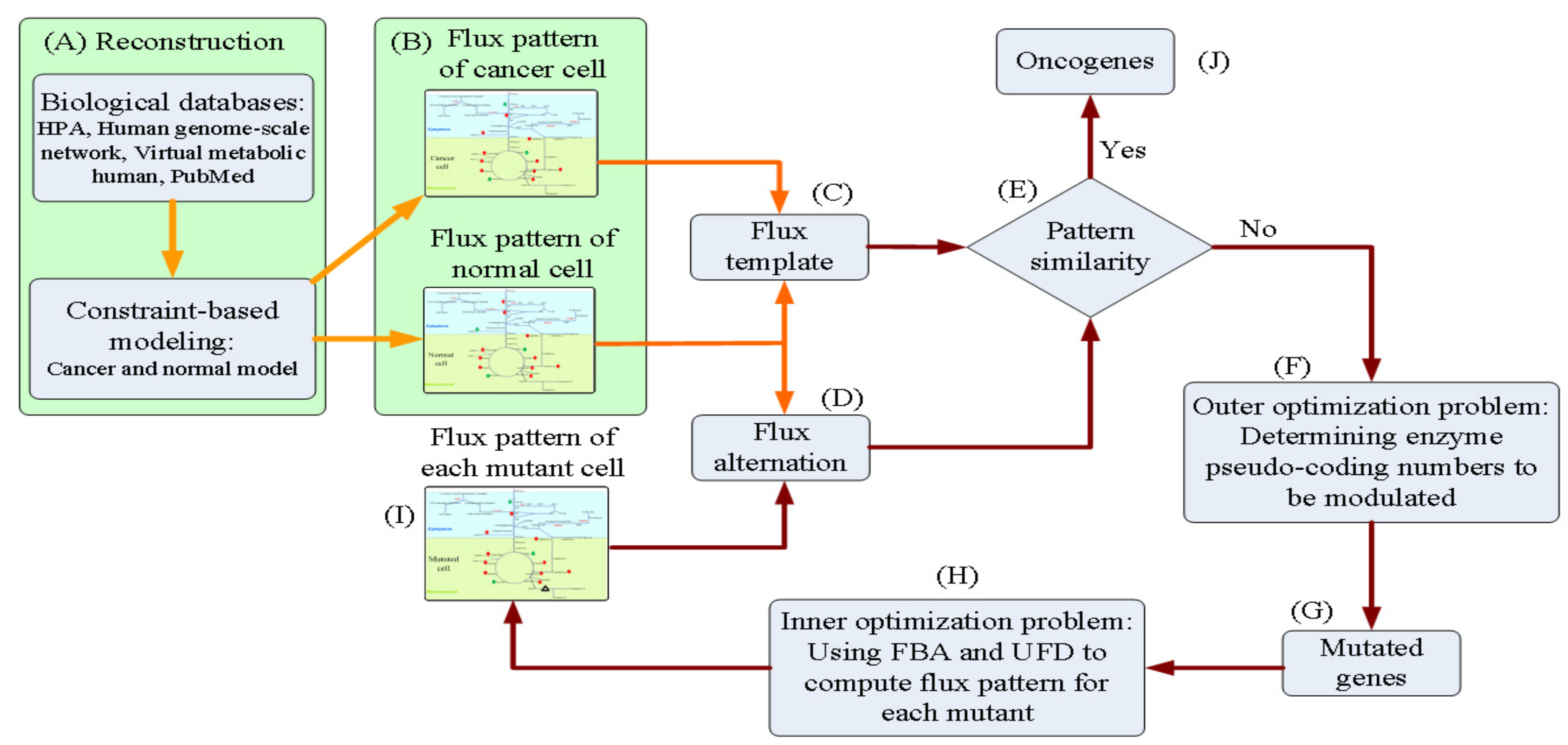
Biology | Free Full-Text | Computer-Aided Design for Identifying Anticancer Targets in Genome-Scale Metabolic Models of Colon Cancer | HTML

Recon3D enables a three-dimensional view of gene variation in human metabolism | Nature Biotechnology

Biomolecules | Free Full-Text | Simultaneous Integration of Gene Expression and Nutrient Availability for Studying the Metabolism of Hepatocellular Carcinoma Cell Lines | HTML

Recon3D enables a three-dimensional view of gene variation in human metabolism | Nature Biotechnology

Adding 3D models to PubChem. Screenshot of the PubChem web page for... | Download Scientific Diagram

OptFill: A Tool for Infeasible Cycle-Free Gapfilling of Stoichiometric Metabolic Models - ScienceDirect
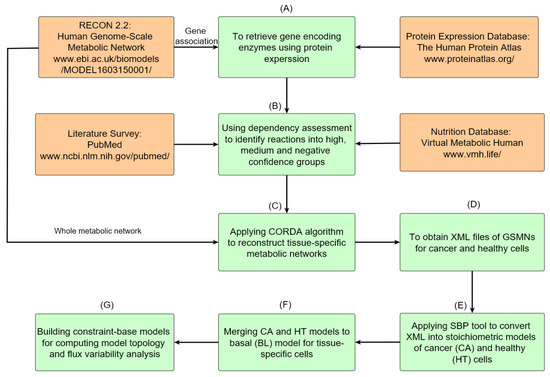
Metabolites | Free Full-Text | Genome-Scale Metabolic Modeling with Protein Expressions of Normal and Cancerous Colorectal Tissues for Oncogene Inference | HTML
Analysis of human metabolism by reducing the complexity of the genome-scale models using redHUMAN | Nature Communications

DIAMOND+MEGAN: Fast and Easy Taxonomic and Functional Analysis of Short and Long Microbiome Sequences - Bağcı - 2021 - Current Protocols - Wiley Online Library

Recon3D enables a three-dimensional view of gene variation in human metabolism | Nature Biotechnology
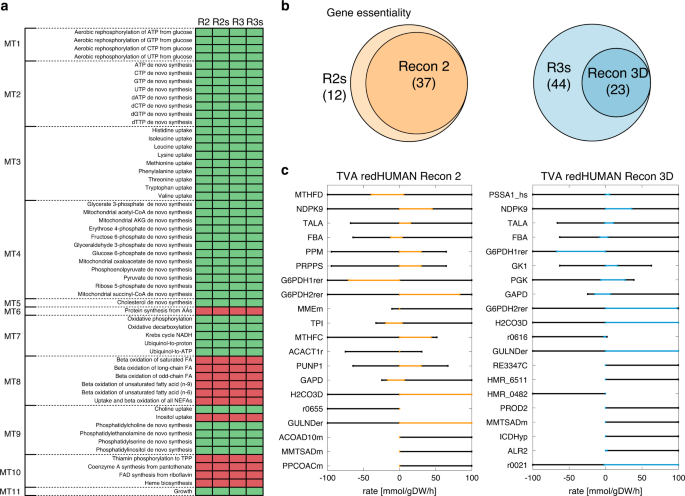
Analysis of human metabolism by reducing the complexity of the genome-scale models using redHUMAN | Nature Communications
Increasing consensus of context-specific metabolic models by integrating data-inferred cell functions

Recon3D enables a three-dimensional view of gene variation in human metabolism | Nature Biotechnology
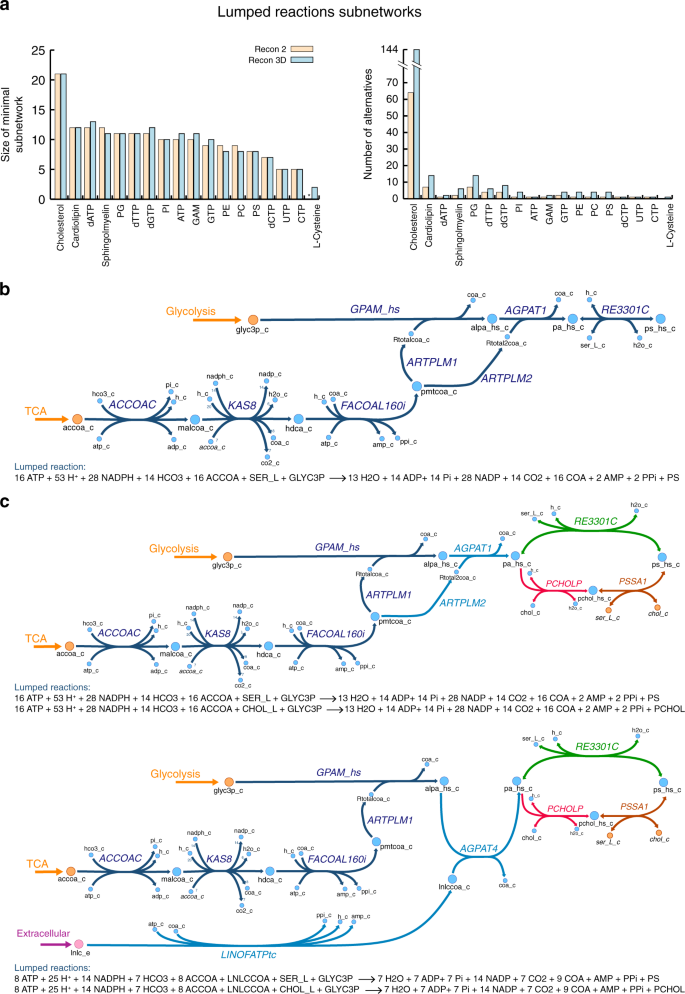
Analysis of human metabolism by reducing the complexity of the genome-scale models using redHUMAN | Nature Communications

Recon3D enables a three-dimensional view of gene variation in human metabolism. - Abstract - Europe PMC
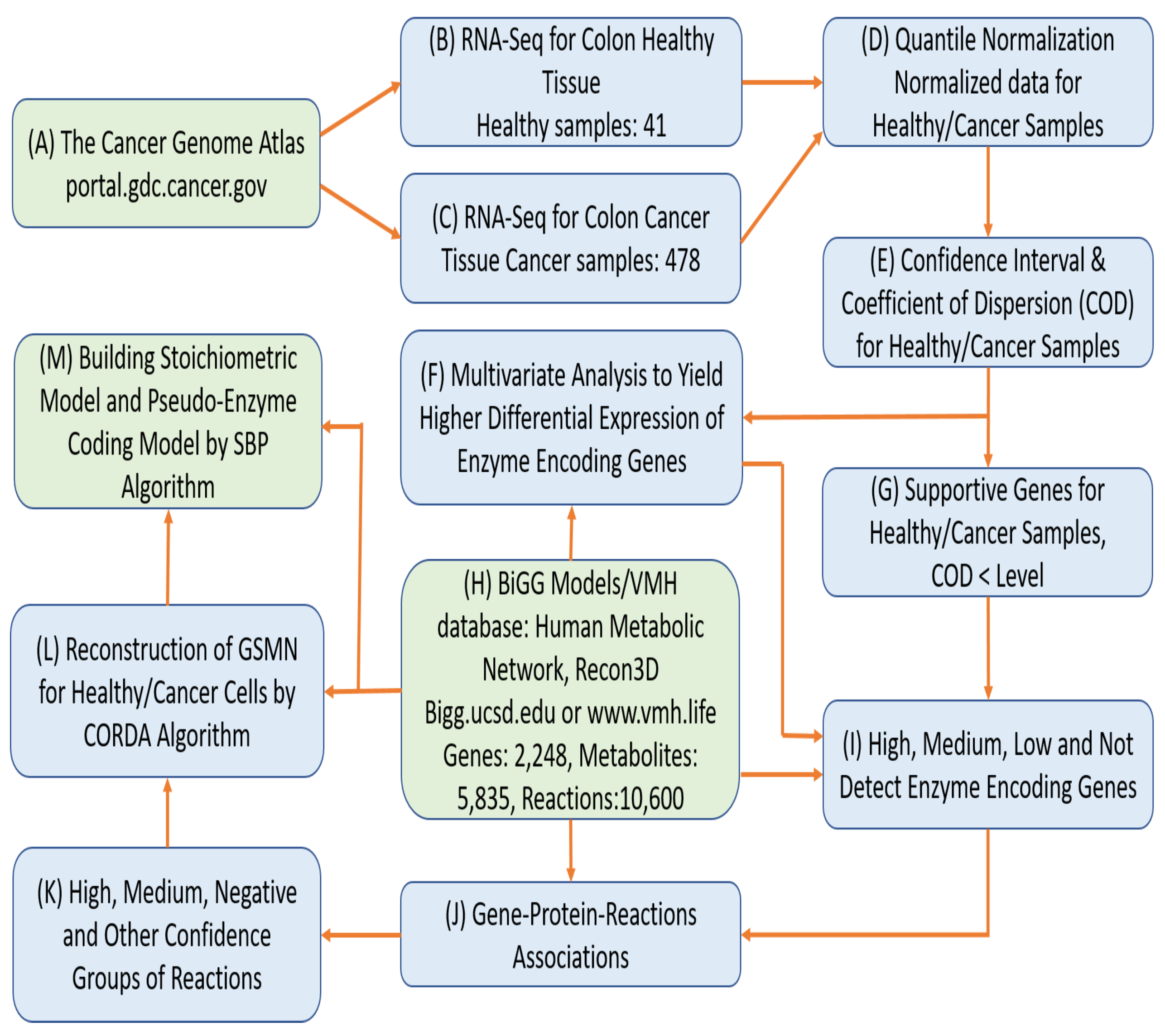
Biology | Free Full-Text | Computer-Aided Design for Identifying Anticancer Targets in Genome-Scale Metabolic Models of Colon Cancer | HTML

Genome‐scale metabolic modeling reveals SARS‐CoV‐2‐induced metabolic changes and antiviral targets | Molecular Systems Biology
PDF) Comparative evaluation of atom mapping algorithms for balanced metabolic reactions: Application to Recon 3D